Présentation de travaux de recherche à la conférence 3DSIG 2016
Des travaux de recherche pour lesquels la société ATRAGENE a participé ont été retenus pour une communication orale à la conférence 3Dsig 2016 Structural Bioinformatics & Computational Biophysics. La présentation a été faite par le Dr Alexandre de Brevern. Le résumé de la présentation, en anglais, se trouve ci-dessous :
Structural properties of frameworks VHHs at the light of a structural alphabet
Introduction
VHHs are interesting biotechnological and biomedical agents due to their size, physico-chemical properties and high thermal stability(1). Currently many VHHs are used in various therapeutic trials and biotechnological applications, for which they need to be modified or humanized. VHHs are derived from classical IgGs, thus have a common structural fold and related properties. They have VH fold composed of constant regions called Frameworks Regions (FRs) with alternating Complementarity Determining Regions (CDRs). The CDRs are highly variable and form the antigenic determinant.
Previously, others and we have noticed that the structural modeling of VHHs is more complicated than expected(2-3). We suspect that this problem arises not only due to the highly flexible conformations of CDRs but also the usage of most related FRs blindly to perform comparative modeling, although they are more structured and conserved than CDRs. Hence we decided to study FRs
precisely.
An important point –at this step- is the usage of a SA, a library of N structural prototypes (the letters). Each prototype is representative of a backbone local structure of 5-residues length. It allows translating a 3D protein structure into a 1D-sequence string (see Figure 1).
It is particularly helpful to analyze protein structures, protein flexibility, molecular dynamics simulations, amino acid equivalence for protein engineering, allostery, binding sites, prediction of protein structure, etc. Our SA called Protein Blocks (PBs)(4) is composed of 16 prototypes and is the most widely used SA till date(5-6).
Methods
The dataset of protein structures represents 114 PDB files, with 160 VHH1 structures. Only one VHH was conserved if they were identical in terms of sequences and structures. The complete dataset of VHH structures is so composed of 133 complete structures.
Various structural analyses have been performed; the most important one focuses on Protein Blocks(4-6). PBs are a set of 16 local prototypes of 5 residues length, clustered based on !, ψ dihedral angles description. They were obtained using an unsupervised classifier similar to Kohonen Maps and Hidden Markov Models. The PBs m and d can be roughly described as prototypes for central α-helix and central β-strand, respectively, while other PB denotes periphery of these secondary structures, and some mainly associated with coils. PB assignment was carried out using PBxplore tool (https://github.com/pierrepo/PBxplore).
Results & Conclusions
This study is the first attempt to analyze precisely the structural diversity of FRs of VHHs. PB analysis has underlined that each of the four FRs was not actually composed of a single structure but by a main structural pattern and many variant patterns. No link between FR length and the percentage of structures corresponding to the patterns was detected. No correlation was found between FR length and number of variant patterns. FR2 is the framework that has the structural pattern representing the largest number of structures (84%), or accordingly lowest number of variant patterns (5). For FRs at the ends FR1 and FR4, the main pattern represents less than 50% of the structures. The longest framework regionFR3 has 10 subunits representing 37% of the structures. Analyses of the amino acid sequences show no direct connection between the local conformational change and amino acid composition. Hence we propose a new representation of FRs and their specific classification in terms of PBs.
These results indicate that long-range interactions affect the local conformation of this constrained topology. These findings have direct consequences on (i) the design of structural models for similar antibodies, (ii) the modifications / optimizations such as humanization and (iii) drug designing studies implying VHHs.
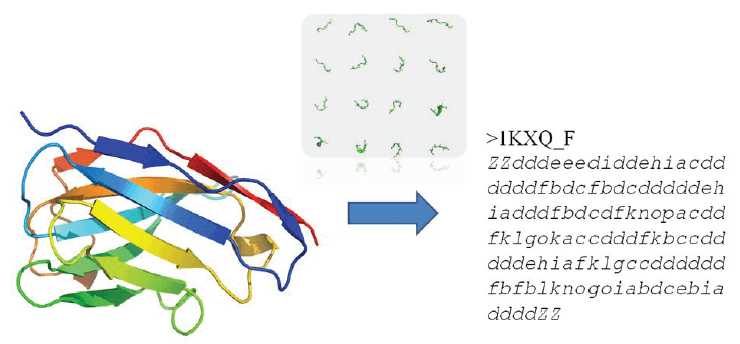
FIGURE 1. Translation of VHH 3D structures in terms of Protein Blocks (PBs). Product is a 1D sequence, like an amino acid sequence, which could be used for several purposes with similar approaches to sequence analyses.
References
1. Hamers-Casterman, C. et al. Nature 363, 446-448 (1993).
2. Smolarek, D. et al. Cell Mol Life Sci 67, 3371-3387 (2010).
3. Sircar, A., Sanni, K.A., Shi. J. & Gray J.J. J Immunol 186, 6357-6367 (2011).
4. de Brevern A.G., Etchebest C. &, Hazout, S. Proteins 41, 271-287 (2000).
5. Joseph A.P. et al. Biophys Rev 2, 137-147 (2010).
6. Craveur P. et al. Frontiers in Molecular Biosciences 2, 20 (2015).
Commentaires
Les commentaires ne sont pas autorisés